The inviscid Burgers’ Equation - A different approach¶
Problem Setup¶
Setting \(u(x,t) = \frac{x}{1+t}\), we expect \(\nu = 0\) as a parameter.
Then \(u_0(x) := u(x,0) = x\).
Using the forward Euler scheme, the equation can be re-written as:
and setting the factor \(u_{n-1} = \mu_{n-1}\) (analogously to the previous subchapter this is the mean of \(u_{n-1}\)) to deal with the non-linearity:
Consider \(u_{n-1}\) to be a Gaussian process.
\(u_{n-1} \sim \mathcal{GP}(0, k_{uu}(x_i, x_j; \theta, l))\)
And the linear operator:
\(\mathcal{L}_x^\nu = \cdot + \tau \nu \frac{d}{dx}\cdot - \tau \mu_{n-1} \frac{d}{dx} \cdot\)
so that
\(\mathcal{L}_x^\nu u_{n-1} = u_n\)
Problem at hand: Estimate \(\nu\) (should be \(\nu = 0\) in the end).
For the sake of simplicity, take \(u := u_{n-1}\) and \(f := u_n\).
Step 1: Simulate data¶
Take data points at \(t = 0\) for \((u_{n-1})\) and \(t = \tau\) for \((u_n)\), where \(\tau\) is the time step.
\(x \in [0, 1], \; t \in \{0, \tau \}\)
In [10]:
tau = 0.001
def get_simulated_data(tau, n=20):
x = np.random.rand(n)
y_u = x
y_f = x/(1+tau)
return (x, y_u, y_f)
(x, y_u, y_f) = get_simulated_data(tau)
In [81]:
show_1(x, y_u, y_f)
Step 2: Evaluate kernels¶
- \(k_{uu}(x_i, x_j; \theta, l) = \theta exp(-\frac{1}{2l}(x_i-x_j)^2)\)
- \(k_{ff}(x_i,x_j;\theta, l,\nu) = \mathcal{L}_{x_i}^\nu \mathcal{L}_{x_j}^\nu k_{uu}(x_i, x_j; \theta, l) \\ = k_{uu} + \tau \nu \frac{d}{dx_i}k_{uu} - \tau \mu_{n-1} \frac{d}{dx_i}k_{uu} + \tau \nu \frac{d}{dx_j}k_{uu} + \tau^2 \nu^2 \frac{d}{dx_i} \frac{d}{dx_j}k_{uu} - \tau^2 \nu \mu_{n-1}\frac{d^2}{dx_i dx_j} k_{uu} - \tau \mu_{n-1} \frac{d}{dx_j}k_{uu} - \tau^2 \nu \mu_{n-1} \frac{d^2}{dx_i dx_j} k_{uu} + \tau^2 \mu_{n-1}^2 \frac{d^2}{dx_i dx_j}k_{uu}\)
- \(k_{fu}(x_i,x_j;\theta,l,\nu) = \mathcal{L}_{x_i}^\nu k_{uu}(x_i, x_j; \theta, l) \\ = k_{uu} + \tau \nu \frac{d}{dx_i}k_{uu} - \tau \mu_{n-1}\frac{d}{dx_i}k_{uu}\)
- \(k_{uf}(x_i,x_j;\theta, l, \nu)\) is given by the transpose of \(k_{fu}(x_i,x_j;\theta, l, \nu)\).
Steps 3 and 4: Compute NLML and optimize the hyperparameters¶
In [76]:
m = minimize(nlml, np.random.rand(3), args=(x, y_u, y_f, 1e-7), method=\
"Nelder-Mead", options = {'maxiter' : 1000})
In [77]:
m.x[2] # This is our inferred value for \nu
Out[77]:
0.00021347647791778476
Step 5: Analysis w.r.t. the number of data points (up to 25):¶
In this section we want to analyze the error of our algorithm and plot its time complexity.
1. Plotting the error in our estimate:
The error is given by \(| \nu_{estimate} - \nu_{true} |\).
We plot the error with respect to the number of data samples for five runs of the program:
In [100]:
show_2(lin, res)
<Figure size 360x216 with 0 Axes>
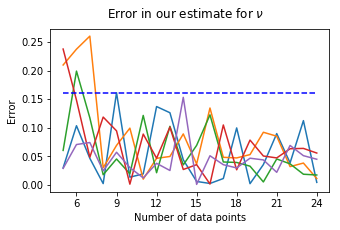
We see that for n sufficiently large (in this case \(n \geq 8\)), we can assume the error to be bounded by 0.16.
2. Plotting the error between the solution and the approximative solution:
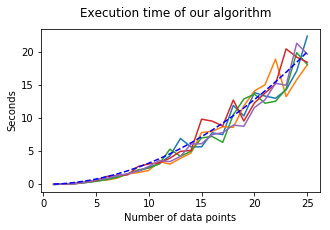
3. Plotting the execution time:
The blue-dashed line follows \(f(x) = 0.032 x^2\).
In [59]:
show_3(lin, timing)
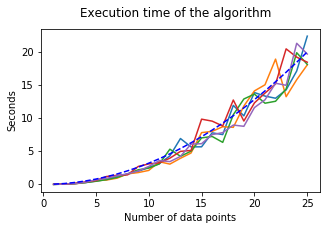
We observe a time complexity of roughly \(\mathcal{O}(n^2)\) (blue-dashed line).
Comparison with a no-mean version¶
\(\tau \nu \frac{d^2}{dx^2}u_{n-1} - \tau x \frac{d}{dx}u_{n-1} + u_{n-1} = u_{n}\)
The linear operator looks just slightly different:
\(\mathcal{L}_x^\nu = \cdot + \tau \nu \frac{d}{dx}\cdot - \tau x \frac{d}{dx} \cdot\)
so that
\(\mathcal{L}_x^\nu u_{n-1} = u_n\).
Our kernels evaluate to:
- \(k_{uu}(x_i, x_j; \theta, l) = \theta exp(-\frac{1}{2l}(x_i-x_j)^2)\)
- \(k_{ff}(x_i,x_j;\theta, l,\nu) = \mathcal{L}_{x_i}^\nu \mathcal{L}_{x_j}^\nu k_{uu}(x_i, x_j; \theta, l) \\ = k_{uu} + \tau \nu \frac{d}{dx_i}k_{uu} - \tau x \frac{d}{dx_i}k_{uu} + \tau \nu \frac{d}{dx_j}k_{uu} + \tau^2 \nu^2 \frac{d}{dx_i} \frac{d}{dx_j}k_{uu} - \tau^2 \nu x\frac{d^2}{dx_i dx_j} k_{uu} - \tau x \frac{d}{dx_j}k_{uu} - \tau^2 \nu x \frac{d^2}{dx_i dx_j} k_{uu} + \tau^2 x^2 \frac{d^2}{dx_i dx_j}k_{uu}\)
- \(k_{fu}(x_i,x_j;\theta,l,\nu) = \mathcal{L}_{x_i}^\nu k_{uu}(x_i, x_j; \theta, l) \\ = k_{uu} + \tau \nu \frac{d}{dx_i}k_{uu} - \tau x\frac{d}{dx_i}k_{uu}\)
- \(k_{uf}(x_i,x_j;\theta, l, \nu)\) is given by the transpose of \(k_{fu}(x_i,x_j;\theta, l, \nu)\).
After constructing the nlml we get:
In [17]:
m = minimize(nlml, np.random.rand(3), args=(x, y_u, y_f, 1e-7), method=\
"Nelder-Mead", options = {'maxiter' : 1000})
m.x[2] # This is our prediction for \nu
Out[17]:
0.00042724953581625225
We can analyze the error in multiple runs and look at the execution time:
1. Plotting the error in our estimate for \(\nu\)
The error is given by \(| \nu_{estimate} - \nu_{true} |\).
In [30]:
show_4(lin, est, res)
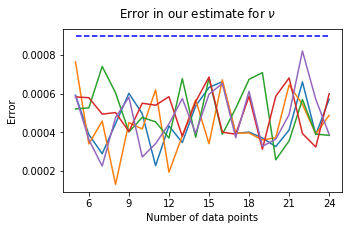
We see that for n sufficiently large (in this case \(n \geq 5\)), we can assume the error to be bounded by 0.0009.
2. Plotting the execution time:
In [57]:
show_5(lin, timing)
The time complexity seems to be as before roughly \(\mathcal{O}(n^2)\).