2D Linear operator with one parameter¶
Along the same lines as the previous example, we take a two-dimensional linear operator corresponding to a two-dimensional system with a single parameter. As previously explained, we set our problem up as follows:
It is easy to verify that \(\mathcal{L}_\bar{x}^\phi\) is linear and continuous. A suitable solution for the operator is
Now, we use the same Gaussian priors as before, except that they are now defined on a two-dimensional space.
The parameter estimation problem for the linear operator described above is to estimate \(\phi\), given \(\{x, y_u, y_f\}\). Notice that we evaluate \(u, f\) on the same set of points because it makes sense from a physics point of view.
Simulate data¶
We use \(\phi = 2\) and try to estimate it.
In [2]:
def get_simulated_data(n, phi):
x = np.random.rand(n,2)
y_u = np.multiply(x[:,0], x[:,1]) - x[:,1]**2
y_f = phi*y_u + x[:,1] - 2
return (x, y_u, y_f)
Evaluate kernels¶
The implementation of the kernels is straightforward as done earlier. Consequently, the corresponding code has been omitted from the report.
- \(k_{uu}(\bar{x}_i, \bar{x}_j; \theta) = \theta e^{(-l_1 (x_{i,1}-x_{j,1})^2 -l_2 (x_{i,2}-x_{j,2})^2)}\)
- \(k_{ff}(\bar{x}_i,\bar{x}_j;\theta,\phi) = \mathcal{L}_{\bar{x}_i}^\phi \mathcal{L}_{\bar{x}_j}^\phi k_{uu}(\bar{x}_i, \bar{x}_j; \theta) \\ = \mathcal{L}_{\bar{x}_i}^\phi \left( \phi k_{uu} + \frac{\partial}{\partial x_{j,1}}k_{uu} + \frac{\partial^2}{\partial^2 x_{j,2}}k_{uu}\right) \\ = \phi^2 k_{uu} + \phi \frac{\partial}{\partial x_{j,1}}k_{uu} + \phi \frac{\partial}{\partial x_{i,1}}k_{uu} \\ \quad + \phi \frac{\partial^2}{\partial^2 x_{j,2}}k_{uu} + \frac{\partial}{\partial x_{i,1}}\frac{\partial}{\partial x_{j,1}}k_{uu} + \phi \frac{\partial^2}{\partial^2 x_{i,2}}k_{uu} \\ \quad + \frac{\partial}{\partial x_{i,1}}\frac{\partial^2}{\partial^2 x_{j,2}}k_{uu} + \frac{\partial^2}{\partial^2 x_{i,2}}\frac{\partial}{\partial x_{j,1}}k_{uu} + \frac{\partial^2}{\partial^2 x_{i,2}}\frac{\partial^2}{\partial^2 x_{j,2}}k_{uu}\)
- \(k_{fu}(\bar{x}_i,\bar{x}_j;\theta,\phi) = \mathcal{L}_{\bar{x}_i}^\phi k_{uu}(\bar{x}_i, \bar{x}_j; \theta) \\ = \phi k_{uu} + \frac{\partial}{\partial x_{i,1}}k_{uu} + \frac{\partial^2}{\partial x_{i,2}^2}k_{uu}\)
In [8]:
(x, yu, yf) = get_simulated_data(10, 2)
nlml((1, 1, 1, 0.69), x, yu, yf, 1e-6)
Out[8]:
10.124377463652147
Optimize hyperparameters¶
In [9]:
nlml_wp = lambda params: nlml(params, x, yu, yf, 1e-7)
m = minimize(nlml_wp, np.random.rand(4), method="Nelder-Mead")
In [11]:
np.exp(m.x)
Out[11]:
array([1.29150445e+06, 4.54133519e-05, 5.27567407e-04, 2.00497047e+00])
The optimized hyperparameters are:
| Parameter | Value |
|---|---|
| \(\theta\) | 1.29150445e+06 |
| \(l_1\) | 4.54133519e-05 |
| \(l_2\) | 5.27567407e-04 |
| \(\phi\) | 2.00497047 |
Analysis¶
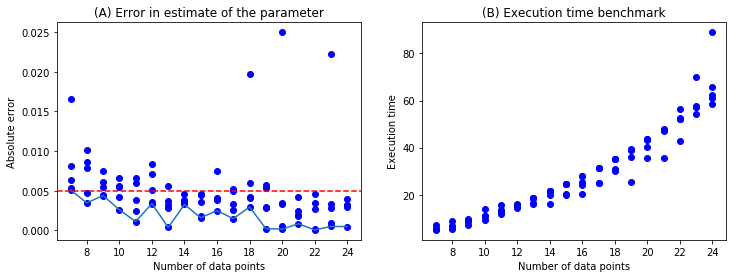
To investigate error properties for this model, we run simulations from 5 to 25 datapoints with 5 samples of random data in each case which is 100 samples. The absolute error in the parameter estimate, \(|\phi_{est} - \phi_{true}|\) is plotted in the left figure below. The execution time is plotted to the right; it includes the time taken to simulate the data and the time taken by the optimization routine.
In [76]:
plt.show()